“Putting the pieces together…”
We are a research group with an open mind towards science, through which we seek to better understand the world around us. Our work is led by Prof. David Friedecký, a scientist and clinical biochemist with over 20 years of experience in a hospital diagnostic laboratory for inborn errors of metabolism and extensive experience in analytical and clinical mass spectrometry. David has been involved in the development of most of the methods used in the laboratory which are also routinely applied for the analysis of samples from patients and newborns with suspected and/or diagnosed inborn errors of metabolism. We are a fellowship of scientists at the Faculty of Medicine and Dentistry, Palacký University Olomouc and University Hospital Olomouc focused on Clinical Mass Spectrometry applications. Our research aims at the utilization of mass spectrometry (mainly combined with liquid chromatography) to perform metabolomic, lipidomic, and multicomponent analyses, which allow detailed insight into understanding the metabolic processes of biological systems. The main research topics cover inborn errors of metabolism, neurodegenerative disorders, cancer, leukemia, and others.

Methods
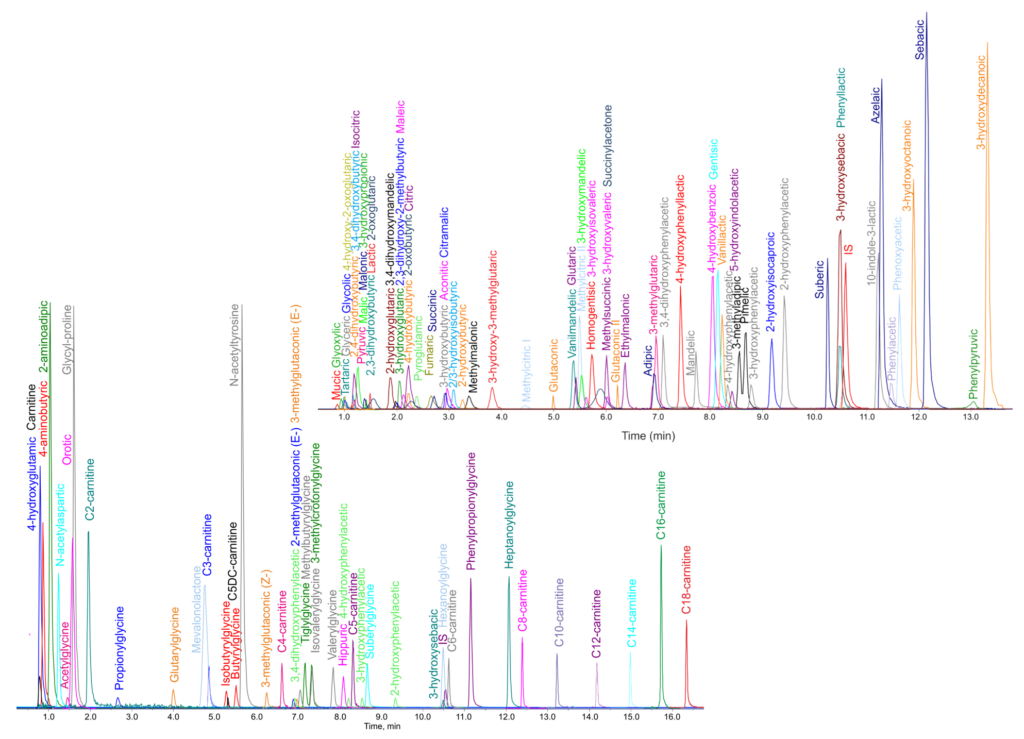
Metabolomics and lipidomics are dynamically evolving fields of analytical biochemistry. We apply both un/targeted approaches for sample analysis. The targeted methods offer relative and or absolute quantitation of selected groups of metabolites and lipids and bring us detailed information about biology; pathobiochemistry; cellular processes and understanding biochemical pathways. We developed a targeted metabolomics method based on HILIC UHPLC/QqQ (6500+ QTrap, Sciex), allowing relative quantitation of 365 compounds in one run (Amino acids | Organic acids | Purine and pyrimidine bases, ribosides and nucleotides | Sugar phosphates | Acylcarnitines | Fatty acids | Carbohydrates | Amines) and targeted lipidomics method for the quantitation of 1000+ lipids in positive and negative mode (RP-UHPLC/QqQ, 6500+ QTrap, Sciex). The untargeted analysis is characterized by the profiling of a broad spectrum of compounds present in the samples. For untargeted profiling and marker discovery, the RP-UHPLC/HR-MS (Orbitrap Elite, Thermo) method is applied.
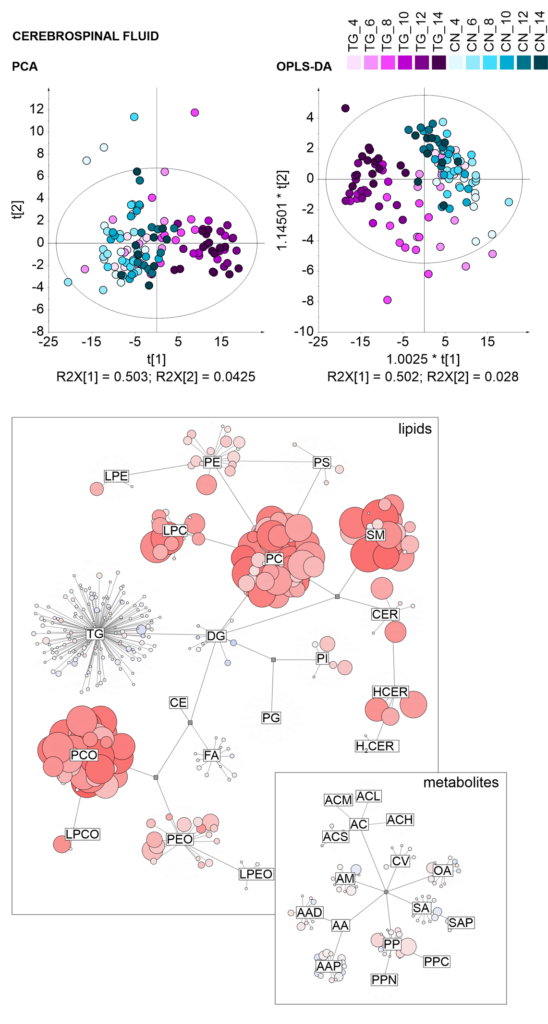
Statistical and graphical tools
- Statistical preprocessing and data evaluation (R software, SIMCA, GraphPad)
- LOESS for QC processing and data transformation (clr, ilr, ln, log, PQN, lnPQN)
- Univariate statistical methods (Boxplots, Volcano plots, testing hypothesis)
- Multivariate data analysis (PCA, Hierarchical Cluster Analysis, PLS-DA, OPLS-DA, Heatmaps, Random forest)
- Correlation analysis, ROC analysis, Power analysis, Z-Score
- New biomarker discovery
- R software, Compound Discoverer (Thermo Fisher Scientific, MA, USA)
- HMDB, SMPDB, Metlin, mzCloud, ChemSpider, PubChem
- Biochemical interpretation/pathway analysis
- KEGG, Lipids maps, Metagene, BRENDA, Reactome